application note sur l'analyse métagénomique du microbiote par séquençage shotgun
document sur l'analyse métagénomique via le séquençage de nouvelle génération
Contenu du document
the swiss dna company
application note · next generation sequencing
metagenomics analysis of microbiota by next generation shotgun sequencing
Understand the genetic potential of your community samples. Provides you with hypothesis-free taxonomic analysis.
introduction
Microbiome studies are often based on the sequencing of specific marker genes, such as the prokaryotic 16S rRNA gene. Such amplicon-based approaches are well established and widely used. However, they have limitations, such as the dependency on a single gene to analyze a whole community, the introduction of PCR bias, and the restriction to describe only the taxonomic composition and diversity. Shotgun metagenomics is a cutting-edge technique for microbiome analysis overcoming said limitations. Whole genomic DNA of a sample is isolated, fragmented, and finally sequenced. This allows a detailed analysis of the taxonomic and functional composition of a microbial community.
microsynth competences and services
Microsynth offers a full shotgun metagenomics service for taxonomic and functional profiling of clinical, environmental, or engineered microbiomes. The service covers the entire process from experimental design, DNA isolation, tailored sequencing to detailed customized bioinformatics analysis. A major advantage of Microsynth’s shotgun metagenomics service is the project-specific consulting and support by our experienced NGS specialists.
experimental design
Microbiome profiling by shotgun metagenomics often means sequencing of numerous samples generating millions or even billions of reads. To gain reliable and high-quality data, a well-planned experimental design including replicates and a clear scientific hypothesis is essential. Our NGS specialists will assist you from the beginning.
dna isolation
Total DNA is either isolated by the customer or this critical step is outsourced. Microsynth has over 15 years of experience in nucleic acid isolation from various demanding matrices such as food, stool, eukaryotic tissues (plants, animals, human), water, or soil.
tailored ngs sequencing
Microsynth will tailor the whole sequencing process to your project requirements, thus providing you with just the right amount of data to answer your questions.
bioinformatics
Taxonomic and functional analysis of metagenomic datasets is challenging. Alignment, binning, and annotation of the large amounts of sequencing reads require expertise and sufficient computational power. Microsynth offers cutting-edge bioinformatics analysis for your shotgun sequencing data using published and well-established methods. Our analysis pipeline will be adjusted to your specific requirements to guarantee scientifically reliable results for your project. After quality processing, the reads are aligned against a protein reference database (e.g., NCBI nr). Taxonomic and functional binning and annotation are performed. The analysis is not restricted to prokaryotes but also includes eukaryotes and viruses. The output comprises all reads, alignments, as well as the taxonomic and functional annotation. Together with the raw and processed data, an HTML report is sent, facilitating the examination, filtering, and sorting of the data.
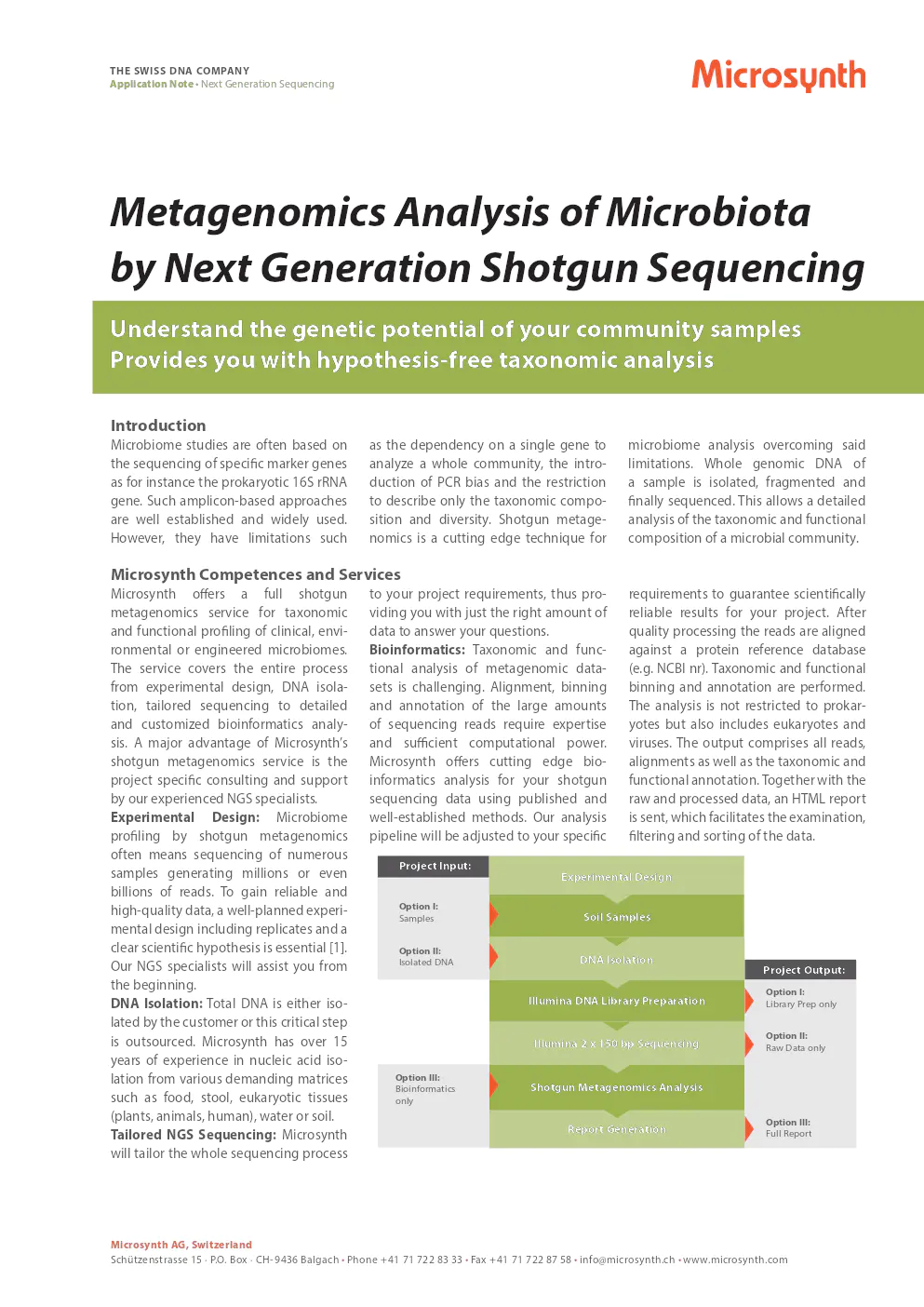
project input
- option i: Samples - Soil Samples
- option ii: Isolated DNA - DNA Isolation
project output
- option i: Library Prep only - Illumina DNA Library Preparation
- option ii: Raw Data only - Illumina 2 x 150 bp Sequencing
- option iii: Full Report - Shotgun Metagenomics Analysis
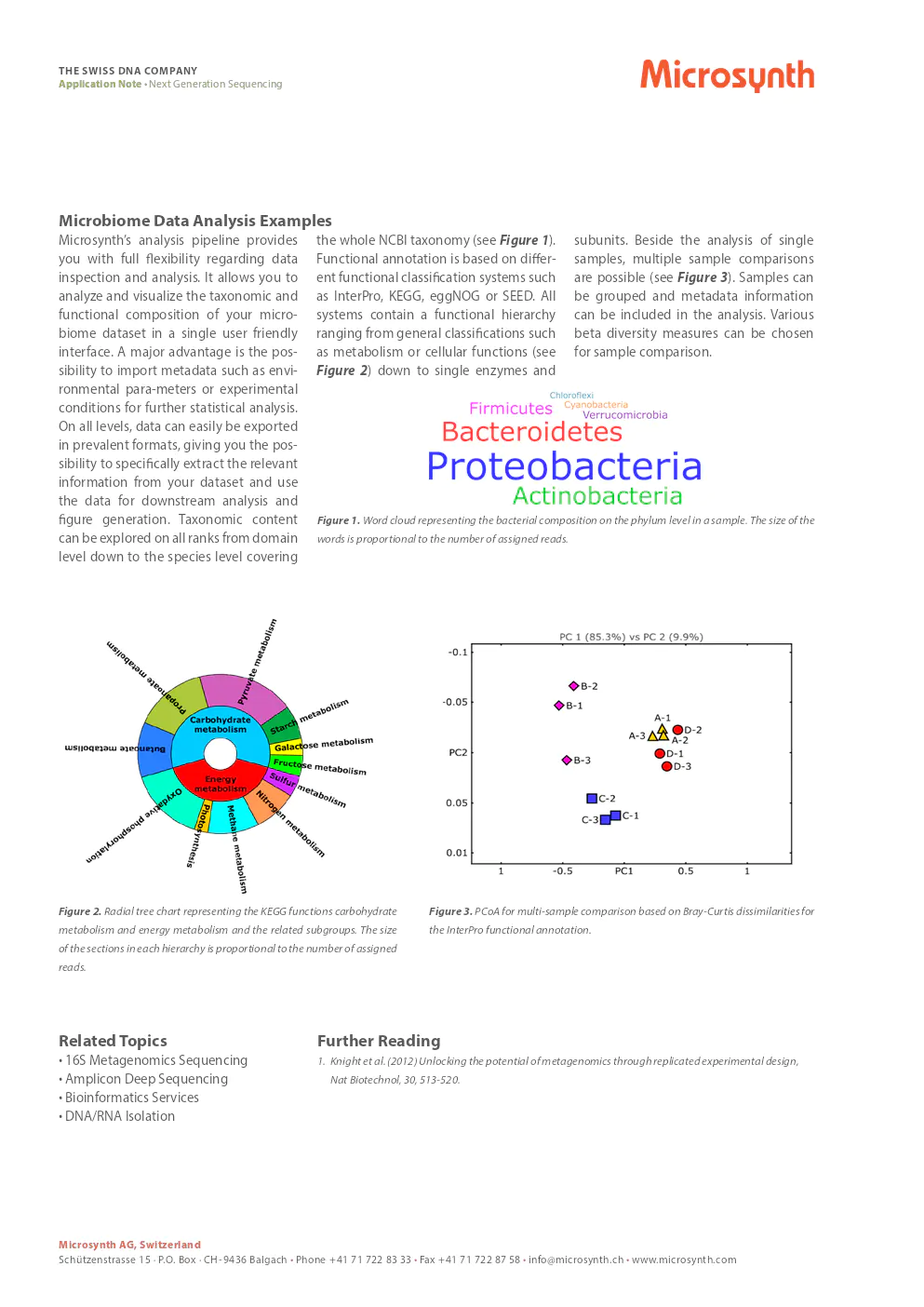
microbiome data analysis examples
Microsynth’s analysis pipeline provides you with full flexibility regarding data inspection and analysis. It allows you to analyze and visualize the taxonomic and functional composition of your microbiome dataset in a single user-friendly interface. A major advantage is the possibility to import metadata such as environmental parameters or experimental conditions for further statistical analysis. On all levels, data can easily be exported in prevalent formats, giving you the possibility to specifically extract the relevant information from your dataset and use the data for downstream analysis and figure generation. Taxonomic content can be explored on all ranks from domain level down to the species level covering the whole NCBI taxonomy. Functional annotation is based on different functional classification systems such as InterPro, KEGG, eggNOG, or SEED. All systems contain a functional hierarchy ranging from general classifications such as metabolism or cellular functions down to single enzymes and subunits. Beside the analysis of single samples, multiple sample comparisons are possible. Samples can be grouped and metadata information can be included in the analysis. Various beta diversity measures can be chosen for sample comparison.
related topics
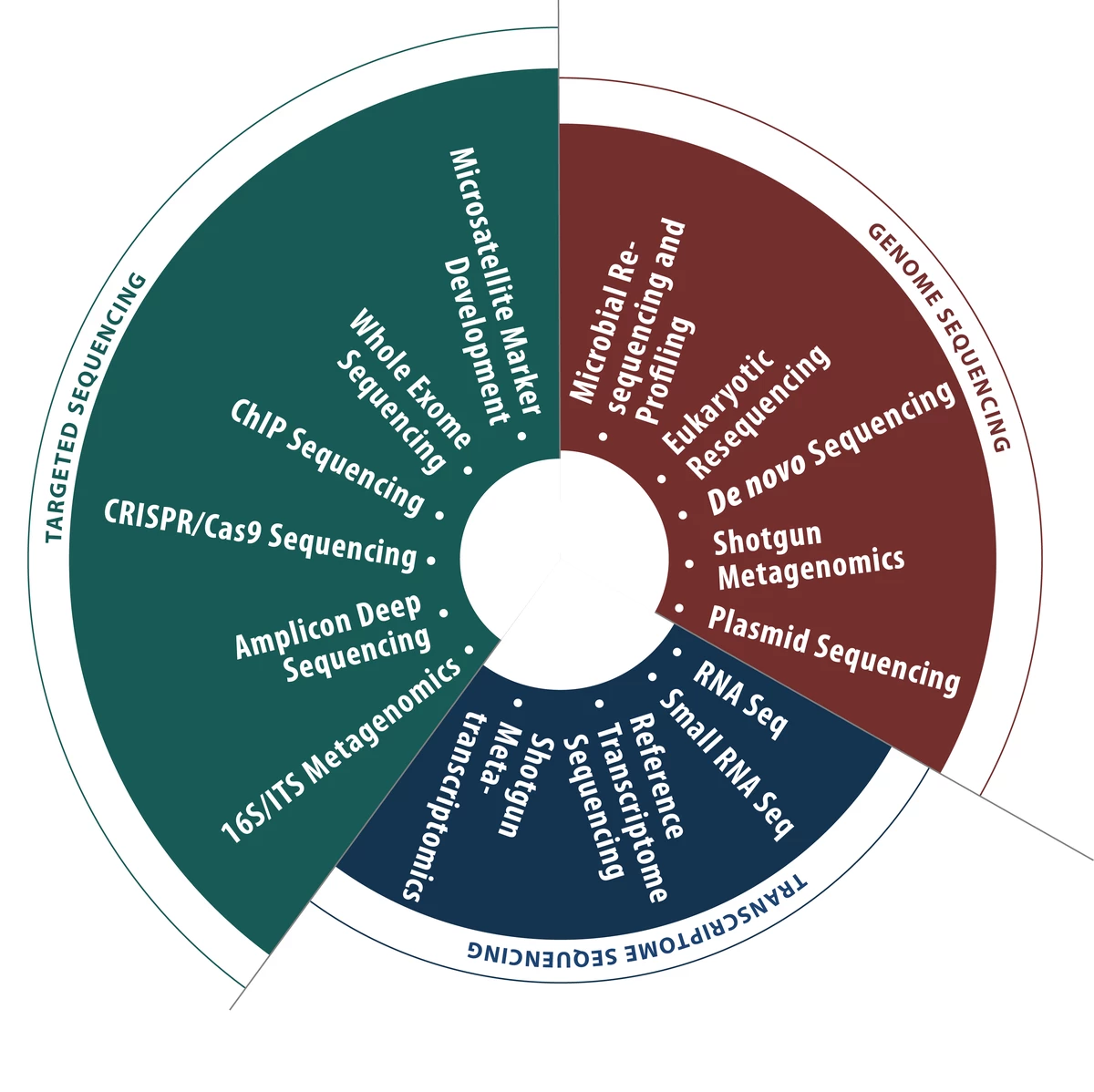
- 16S Metagenomics Sequencing
- Amplicon Deep Sequencing
- Bioinformatics Services
- DNA/RNA Isolation
microsynth ag, switzerland
Schützenstrasse 15 · P.O. Box · CH?-?9436 Balgach · Phone +?41 71 722 83?33 · Fax +?41 71 722 87?58 · info@microsynth.ch · www.microsynth.com