livre blanc sur le séquençage de nouvelle génération par microsynth
document sur la planification et l'exécution des expériences RNA-Seq

Contenu du document
RNA sequencing
An introduction to efficient planning and execution of RNA sequencing (RNA-Seq) experiments.
Motivation for RNA Sequencing
RNA sequencing (RNA-Seq) refers to a method based on next generation sequencing (NGS) technologies to study transcriptomes. Unlike microarray-based technologies that depend on measuring hybridization intensities to predesigned probes, RNA-Seq relies on discrete counting of sequenced molecules, requiring no prior genome knowledge and is hypothesis-free. It has a higher dynamic range compared to microarray experiments and due to falling costs of NGS technologies, RNA-Seq has become the gold standard in transcriptome studies, capable of detecting novel isoforms, alternative splice sites, rare transcripts, gene-fusions, and non-coding transcripts in a single experiment.
Experimental Design
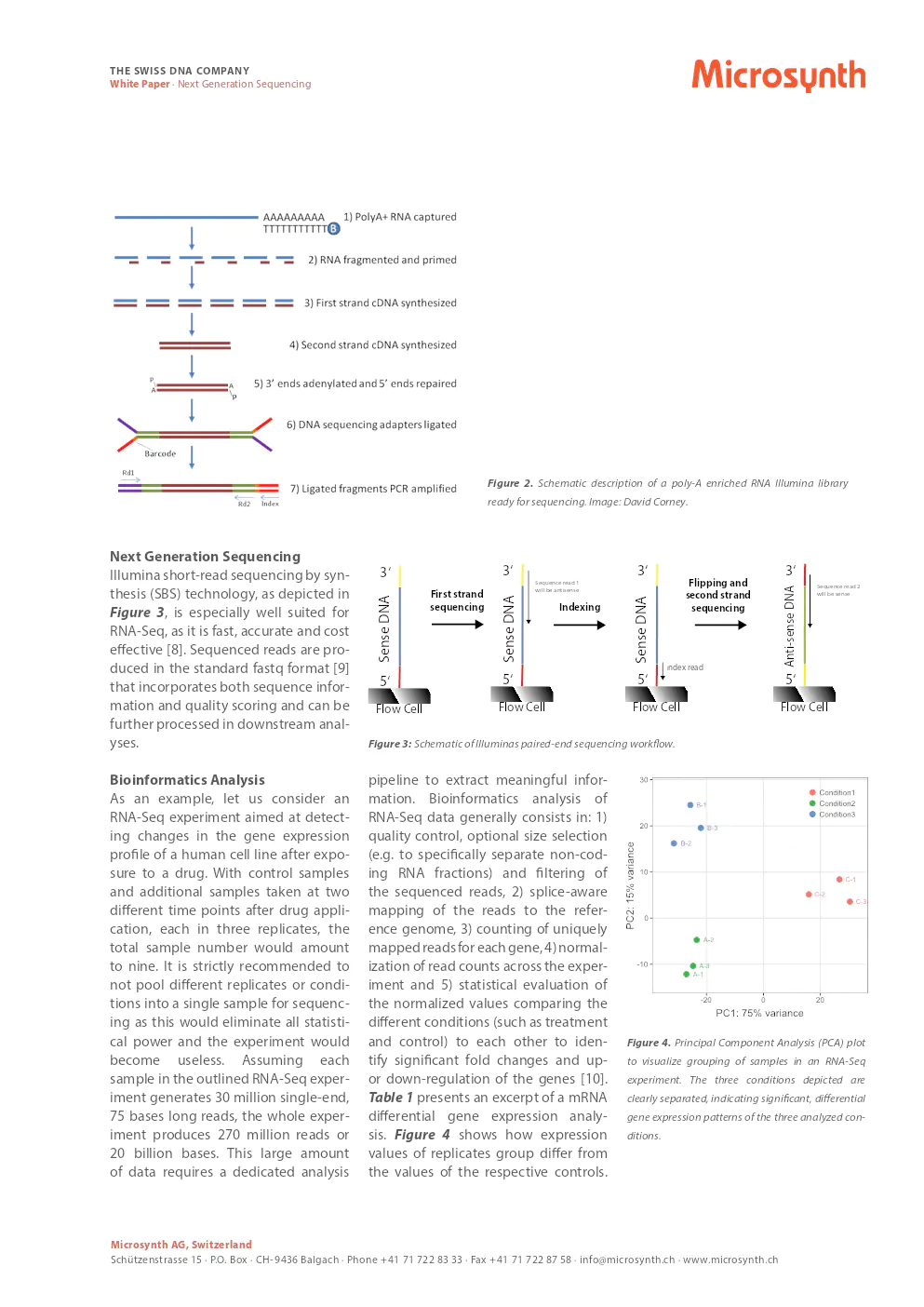
A generalized workflow of a typical RNA-Seq study involves several steps including sample collection, total RNA isolation, RNA library preparation, Illumina short read sequencing, and data analysis which includes report generation.
- Samples: Sample collection must be carefully designed considering experimental conditions like drug treatments and controls to determine transcript abundance differences.
- Total RNA Isolation: High quality RNA is essential as degradation may introduce biases, affecting the transcript profile.
- RNA Library Preparation: Depending on the RNA type and organism, different library construction methods are used, such as polyA enrichment for eukaryotes or ribosomal RNA depletion for prokaryotes.
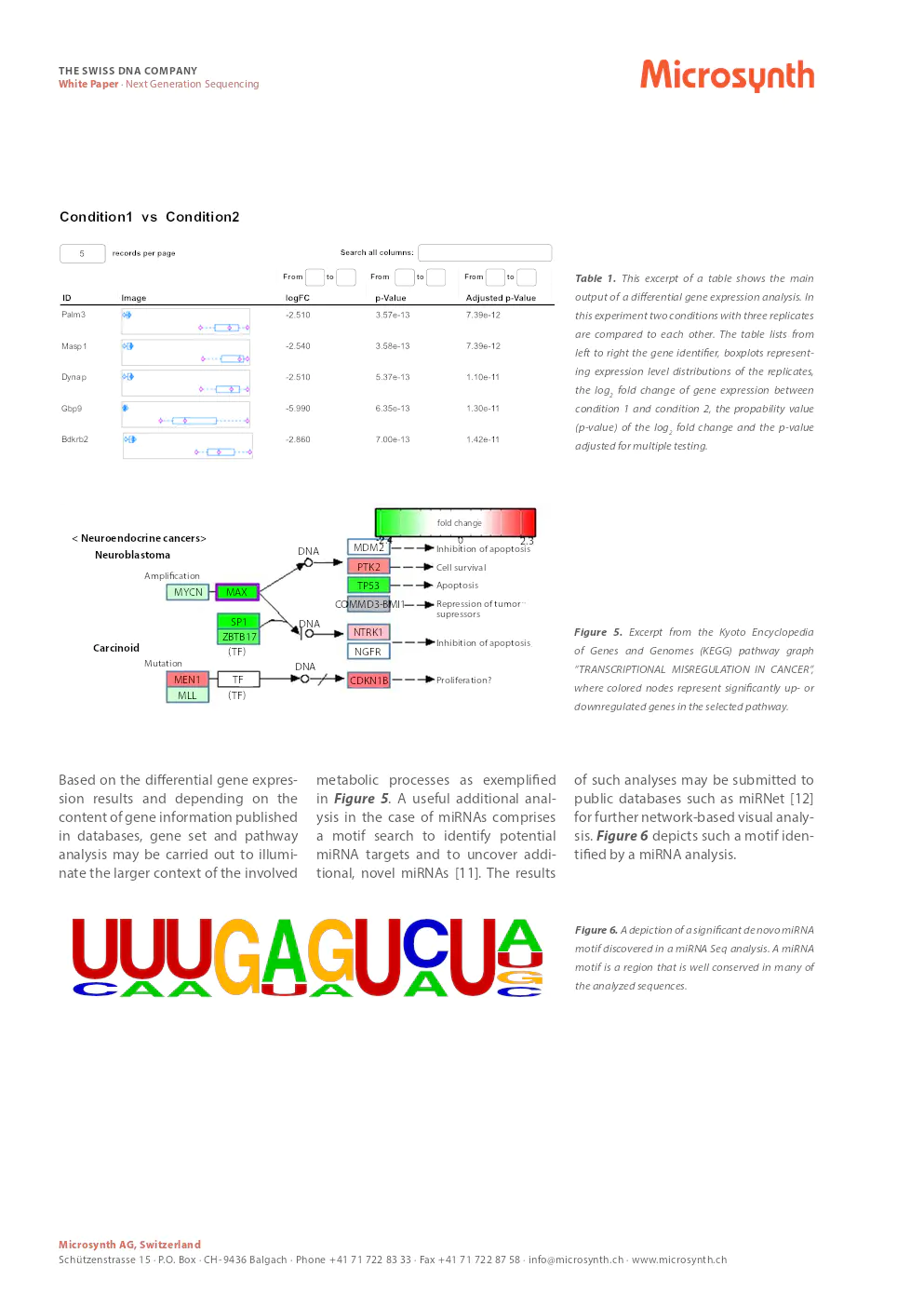
- Sequencing: Illumina short-read sequencing by synthesis is recommended for its speed, accuracy, and cost-effectiveness.
- Data Analysis: Bioinformatics analysis typically involves quality control, size selection, filtering, splice-aware mapping, gene count normalization, and statistical evaluation of normalized values.
Sequencing and Differential Expression Analysis of Coding and Non-coding RNA
Differential expression analysis of RNA-Seq data involves comparison of datasets from experimental conditions to controls, focusing on messenger RNA (mRNA) and non-coding miRNAs, which can be used to develop biomarkers specific to medical conditions.
Bioinformatics Analysis
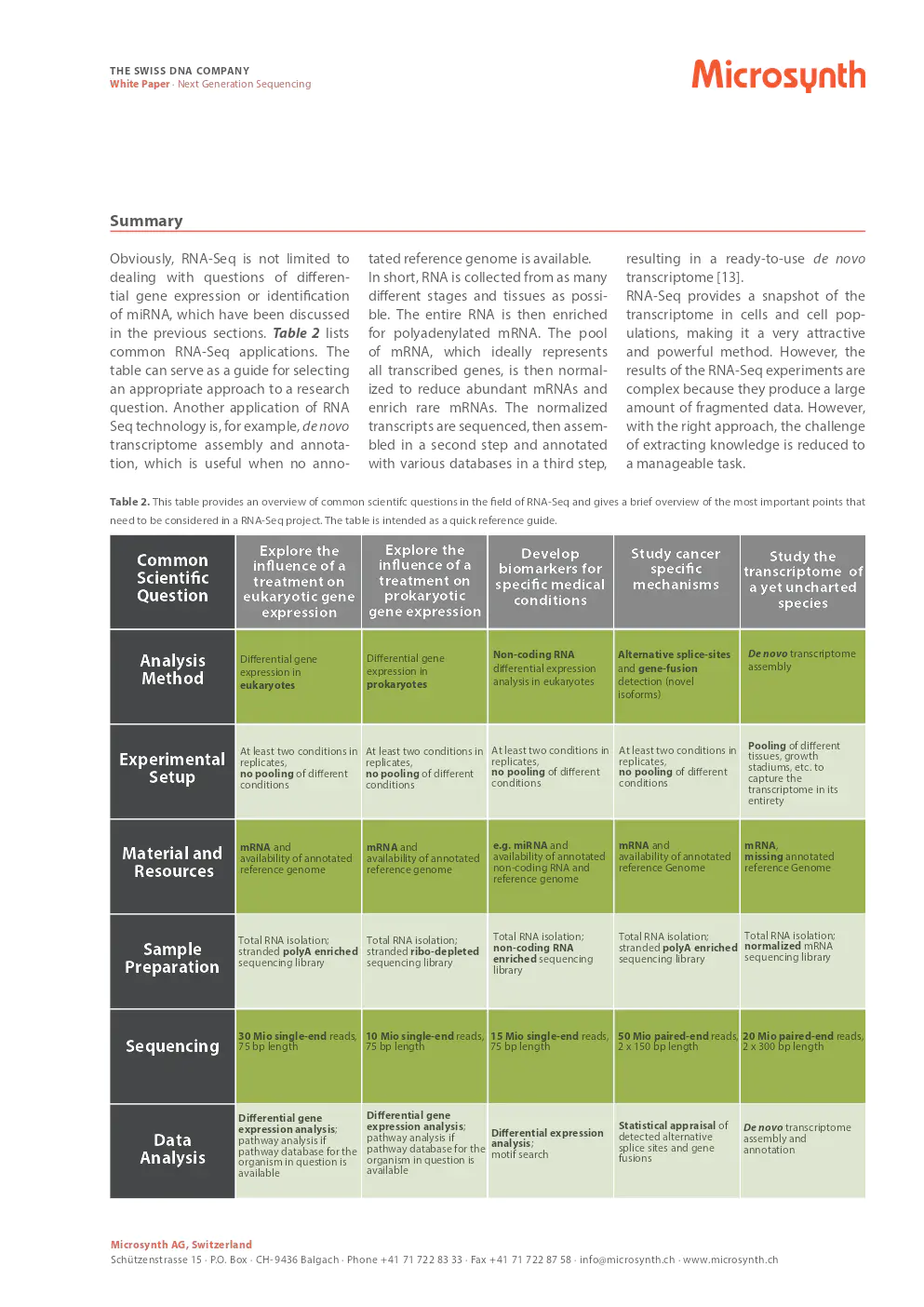
An example of an RNA-Seq experiment involves detecting gene expression changes after drug exposure, where different conditions are pooled for sequencing. Analysis includes quality control, mapping, counting, normalization, and statistical comparison of conditions.
Summary
RNA-Seq is a powerful method providing a snapshot of the transcriptome, dealing with differential gene expression, miRNA analysis, transcriptome assembly, and annotation. It's essential in exploring eukaryotic gene expression, developing biomarkers, and understanding uncharted species' transcriptomes.
References
This white paper cites numerous studies and references to support the findings and methodologies discussed, providing a comprehensive overview of RNA-Seq applications and insights.